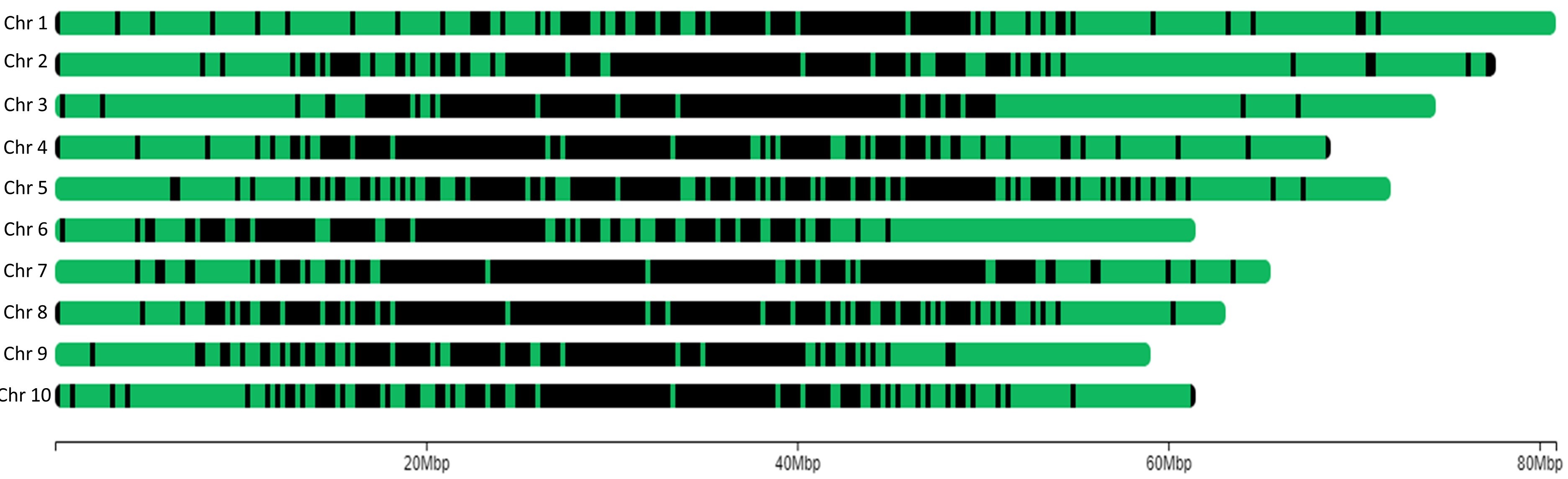
Pearl Millet mid-density genotyping services
The pearl millet SNP mid-density genotyping panel is comprised of 3501 markers distributed across all the 7 chromosomes, including 5 that were derived from an unanchored scaffold, designed on the 843B Cenchrus americanus reference genome (figshare.com/ndownloader/files/37686405). Over 63% of the SNPs (2218) are within genic regions, of which 613 are in coding regions spread over 2096 genes, showing good functional potential.