The CIMMYT Global Wheat Program currently optimizes, and pilots accelerated breeding schemes to increase overall genetic gains through the ‘Accelerating Genetic Gains in Maize and Wheat’ and ‘Zinc mainstreaming in Wheat’ projects funded by the BMGF, FCDO, USAID, and FFAR. A robust, cost-effective mid-density SNP platform is ideal for this type of genomic-assisted breeding approach.
The DArTAG wheat panel has been updated during 2021 and currently contains 3900 SNP markers distributed across the 21 bread wheat chromosomes (Figure 1). Sequences included in this panel are mainly derived from CIMMYT genomics resources: a large genotyping-by-sequencing (GBS) datasets on more than 60K CIMMYT elite breeding lines and a microarray dataset (90K iSelect SNP genotyping array) on more than 1000 diverse wheat accessions (Sehgal et al. 2020, Velu et al. 2016).
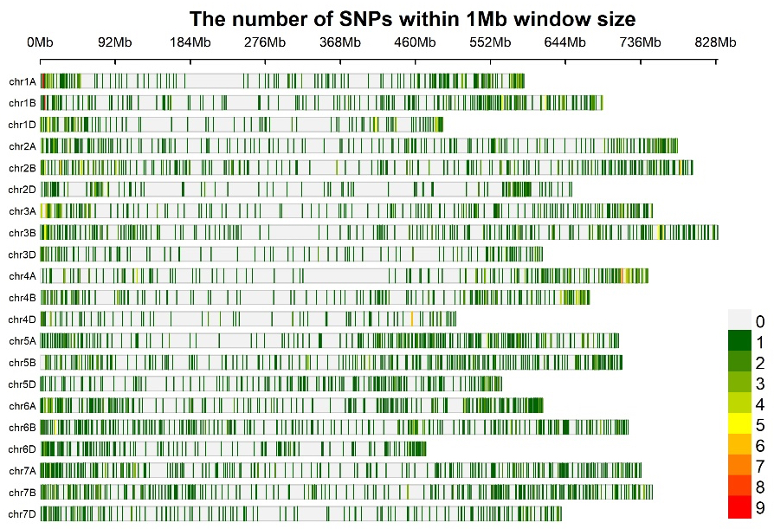
Figure 1. SNP density plot for 3900 SNPs in the current Wheat DArTAG panel (Source: Susanne Dreisigacker)
The SNP markers were selected based on their minor allele frequency, genome and chromosome allocation, haplotypes formed in CIMMYT elite germplasm and an appropriate overall genome distribution. In this updated panel, 2022 sequences remain from the initial panel design, and 1878 sequences were added to cover gaps, additional associated markers to genes and QTL. A summary is given in table 1.
The panel links to a few publicly available genotyping platforms: GBS, the exome capture wheat sequencing panel contributed by Kansas State University (He et al. 2019) and the 35K Axiom Breeders array contributed by the University of Bristol. A total of 110 quality control (QC) and156 associated markers for several key traits i.e., disease resistance, quality, and agronomic traits. Low-density genotyping for some of these traits is utilized in CIMMYT wheat breeding programs (please refer to the KASP marker list).
This panel is suitable for genetic diversity studies and genomic selection (GS) application.
A detailed list of markers is available here (Excel download).
Table 1. Number of sequences selected for DArTAG panels
| Type | # of sequences |
| Gene-based SNPs/Indels | 156 |
| QTL - associated SNPs | 312 |
| QC candidate SNP | 110 |
|
Genome-wide SNPs |
1802 - GBS-derived: 1280 - 90K iSelect array-derived: 522 |
| Exome capture SNPs (KSU) | 500 |
| UK35K Axiom Array SNPs (Bristol) | 1005 |
| Total | 3900 |
References
- He, F., Pasam, R., Shi, F. et al. (2019). Exome sequencing highlights the role of wild-relative introgression in shaping the adaptive landscape of the wheat genome. Nat. Genet 51, 896–904. https://doi.org/10.1038/s41588-019-0382-2.
- Sehgal, D., Mondal, S., Crespo-Herrera, L. et al. (2020). Haplotype-Based, Genome-Wide Association Study Reveals Stable Genomic Regions for Grain Yield in CIMMYT Spring Bread Wheat. Front. Genet., 11, 1427. http://doi.rog/10.3389/fgene.2020.589490.
- Velu, G., Crossa, J., Singh, R.P. et al. (2016). Genomic prediction for grain zinc and iron concentrations in spring wheat. Theor. Appl. Genet. 129(8), 1595-1605. https://doi.org/10.1007/s00122-016-2726-y.

