Groundnut (Arachis hypogaea L.), also known as peanut, is cultivated over 34.1 million hectares with an annual production of 66.3 million tonnes (FAOSTAT, 2020). The availability of reference genomes for both the subspecies (A. hypogaea ssp fastigiata and A. hypogaea ssp hypogaea) of cultivated groundnut in 2019 have boosted more precise genomic studies and higher integration of genomic tools in the modern breeding programs across the world (Pandey et al. 2020).
This cost-effective mid-density SNP panel containing 2500 SNP markers with an average density of about 1 SNP per Mbp distributed across 20 chromosomes provides a powerful tool to groundnut breeders across the world.
The detailed list of markers is available here.
The SNPs have been identified using the whole genome re-sequencing of 263 cultivated accessions of groundnut reference. This diverse set has representative genotypes from all the four major agronomic types, namely Spanish Bunch, Valencia Bunch, Virginia Bunch and Virginia Runner from different regions such as South America, North America, East Southern Africa, West Central Africa, South Asia and South East Asia. The sequence alignment was performed with the high-quality reference genome for cultivated groundnut genotype “Tifrunner” (Bertioli et al. 2019; PRJNA419393, Peanutbase).
The high quality 6000 SNPs were filtered after rigorous processes such as identifying linkage disequilibrium (LD) blocks on individual chromosomes, identifying most informative blocks, identifying most informative markers within block and also performing random sampling proportional to size to include 50% markers from each block and added singletons. The marker panel includes 20-quality control (QC) and 72 associated markers for 8 key traits, namely leaf rust resistance, late leaf spot resistance, net blotch resistance, high oleic acid, seed weight, shelling percentage, fresh seed dormancy and blanchability. Low-density genotyping for some of these traits is currently utilised in groundnut breeding programs.
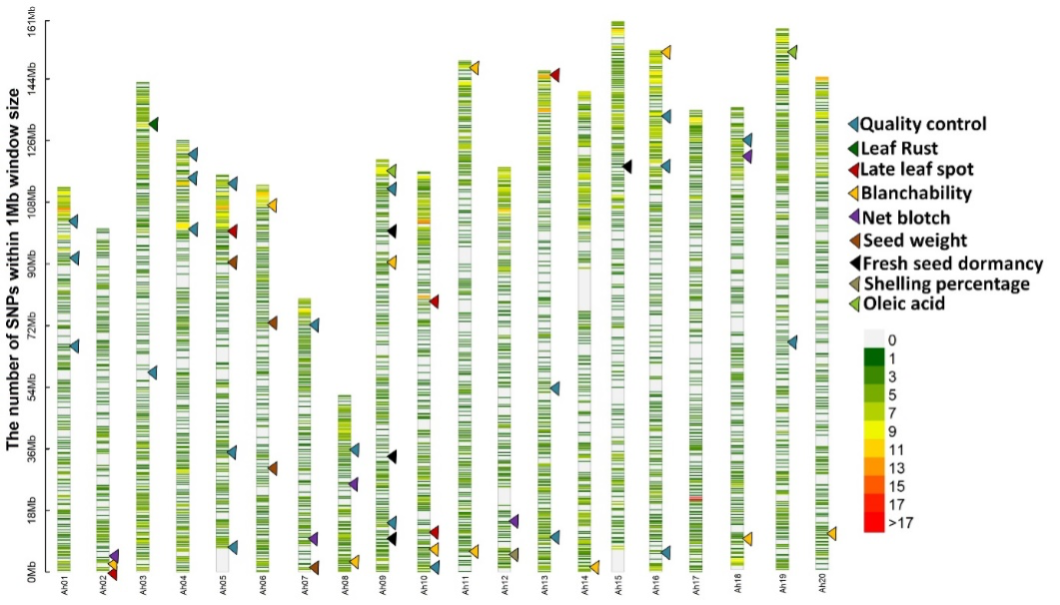
Figure 1. SNP density plot for 2500 SNPs in the Groundnut DArTag SNP panel (Source: Manish Pandey)
This marker panel is suitable for estimating background genome recovery in marker-assisted backcrossing, genomic selection, diversity analyses, and to study genetic purity for efficient germplasm management.
References:
- FAOSTAT (2020). Rome: FAO. http://www.fao.org/faostat/en (accessed on 25 April 2021).
- Bertioli, D.J., Jenkins, J., Clevenger, J. et al. (2019). The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat Genet, 51(5), 877–884. https://doi.org/10.1038/s41588-019-0405-z
- Pandey, M.K., Pandey, A.K., Kumar, R. et al. (2020). Translational genomics for achieving higher genetic gains in groundnut. Theor Appl. Genet 133(5), 1679–1702. https://doi.org/10.1007/s00122-020-03592-2

