Genomics interventions in pigeonpea during the last decade, in particular the availability of a reference genome (Varshney et al. 2012) and whole genome sequence data on multiple lines, have enhanced the level of crop improvement(Varshney et al. 2017; Saxena et al. 2021). Cost-effective genotyping platforms are required to deploy genomics assisted breeding in routine crop improvement programs. Therefore, a mid-density genotyping platform has been developed targeting pigeonpea.
The ICRISAT mid-density array contains 2000 SNPs, precisely selected from large amount of sequencing / genotyping data generated previously in pigeonpea. These selected SNPs will provide the powerful tool to pigeonpea improvement programs.
These SNPs have been identified using the whole genome sequencing / genotyping data on ~450 cultivated accessions. The high quality 8500 SNPs were detected after stringent filtering process such as, identifying linkage disequilibrium (LD) blocks on individual chromosomes, identifying most informative blocks, identifying most informative markers within, etc. (Figure 1). The pigeonpea mid-density genotyping platform contains 2,000 SNPs including quality control (QC) markers and traits associated markers for key traits. The associated traits include resistance to fusarium wilt and sterility mosaic diseases, hybrid purity testing, fertility restoration, etc.
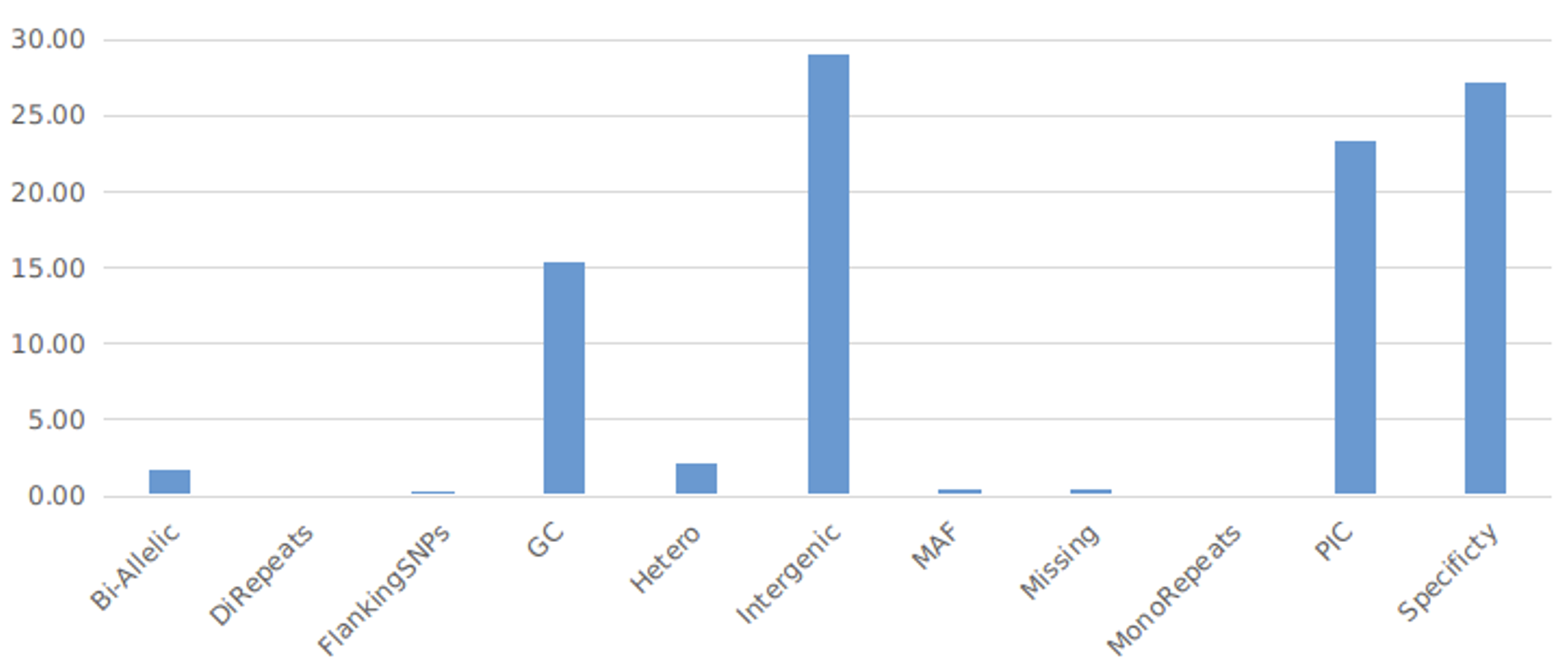
Figure 1. Percent SNPs filtered from the total SNPs used for analysis to develop the Pigeonpea mid-density genotyping panel (Source: Rachit K Saxena).
References
- Saxena RK, Jiang Y, Khan A, Zhao Y, Singh VK, Bohra A, Sonappa M, Rathore A, Sameer Kumar CV, Saxena KB, Reif J, Varshney RK (2021) Characterization of heterosis and genomic prediction based establishment of heterotic pattern for developing better hybrids in pigeonpea. The Plant Genome DOI: 10.1002/tpg2.20125
- Varshney RK, Saxena RK, Upadhyaya HD, Khan AW, Yu Y, Kim C, Rathore A, Kim D, Kim J, An S, Kumar V, Anuradha G, Yamini KN, Zhang W, Muniswamy S, Kim JS, Penmetsa RV, von Wettberg E, Datta SK (2017) Whole-genome resequencing of 292 pigeonpea accessions identifies genomic regions associated with domestication and agronomic traits. Nature Genetics 49: 1082–1088.
- Varshney RK, Chen W, Li Y, Bharti AK, Saxena RK, Schlueter JA, Donoghue MTA, Azam S, Fan G, Whaley AM, Farmer AD, Sheridan J, Iwata A, Tuteja R, Penmetsa RV, Wu W, Upadhyaya HD, Yang SP, Shah T, Saxena KB, Michael T, McCombie WR, Yang B, Zhang G, Yang H, Wang J, Spillane C, Cook DR, May GD, Xu X, Jackson SA (2012) Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nature Biotechnology 30: 83-89.

