The 1k RiCA panel for rice is the mid-density genotyping panel developed at the International Rice Research Institute (IRRI) to enable affordable, efficient genomic selection. It has been implemented at several service providers and platforms, but currently is offered at Agriplex and DArT. Both platforms use custom amplicon workflows that reliably deliver almost all markers from all samples.
The panel consists of roughly 800 genome-wide markers designed to give a fingerprint of breeding material that can be used as the basis for calculating genomics-estimated breeding values in standard GS workflows. The genome-wide panel is supplemented with 22 quality-control markers that are commonly used in purity and F1 hybridity testing at Intertek, providing linkage between datasets, and up to 200 trait markers distinguishing 87 high-value genes and QTLs. Together these capabilities enable genomic selection (including integration of major genes as fixed effects), variety typing, QTL mapping and QTL profiling (fingerprinting) of elite material.
Genome-wide coverage
The panel of genome-wide markers was chosen originally from a panel of c. 4700 SNPs present on an Illumina Infinium panel (Arbalaez et al., 2019), with most informative markers chosen and supplemented with information from the 3000 rice genome sequencing project. Designed primarily to distinguish the indica 1B group, it contains most of the modern breeding material for tropical regions.
Agriplex and DArT panels have 797 and 504 genome-wide markers (Figure 1), respectively and the assessment of polymorphism rates in elite indica material had shown an average of 375 markers polymorphic between any two randomly chosen indica parents (Figure 2). The number of polymorphic markers in japonica material is about of only 180 between most accessions. Yet, modelling has shown that this is sufficient to enable genomic selection and the panel is being used for this purpose in the IRRI breeding programs.
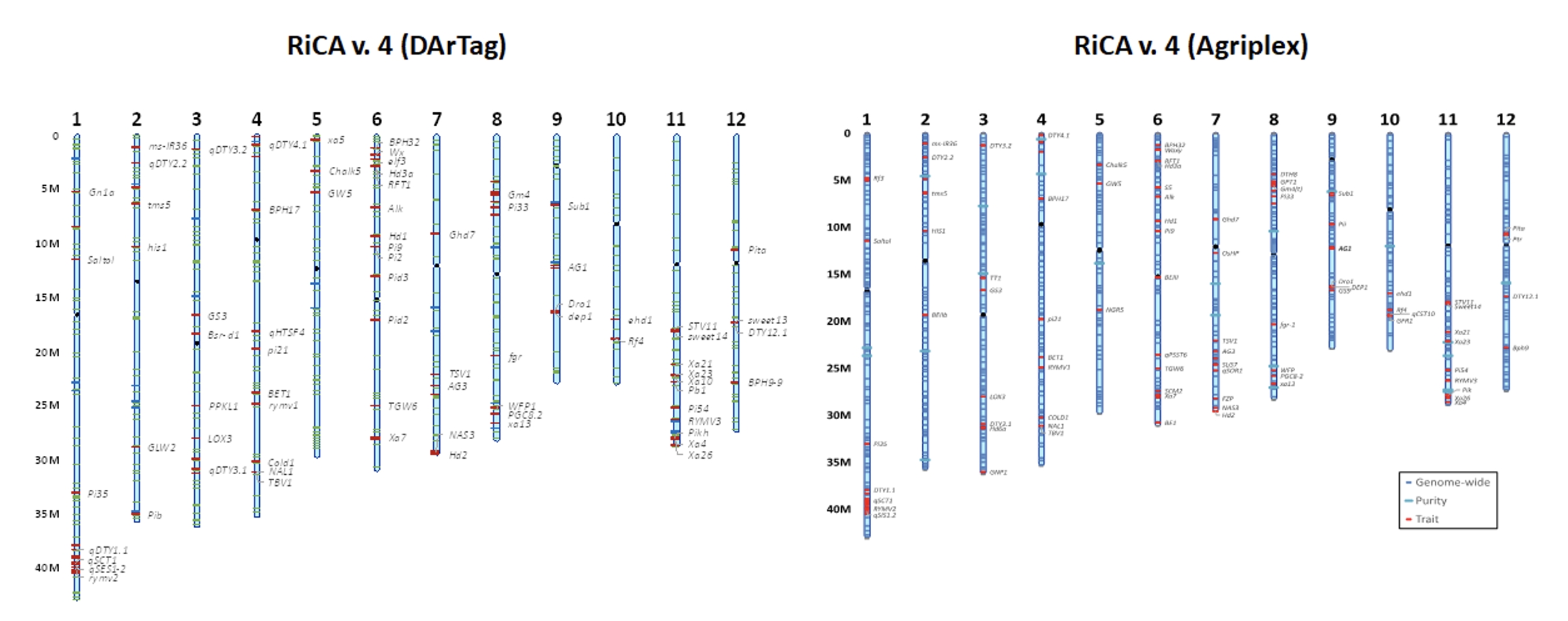
Figure 1. SNP marker position in both existing RiCA v.4 panels
Purity and Trait SNPs incorporated in the panel
The panel of 22 QC SNPs implemented at Intertek, and usually used for parental purity verification and F1 hybridity are also included in the 1k RiCA panel, providing continuity in datasets between the different platforms. Currently, there are 205 trait markers on v. 4 (Agriplex) that distinguish 237 alleles across 87 distinct genes and QTLs targeting genes contributing to maturity, blast, BLB, RYMV, BPH resistance, and many other pest, disease, abiotic stress and grain quality loci. These markers are equally informative across O. sativa background, therefore, with no need of prior information about the material to be fingerprinted.
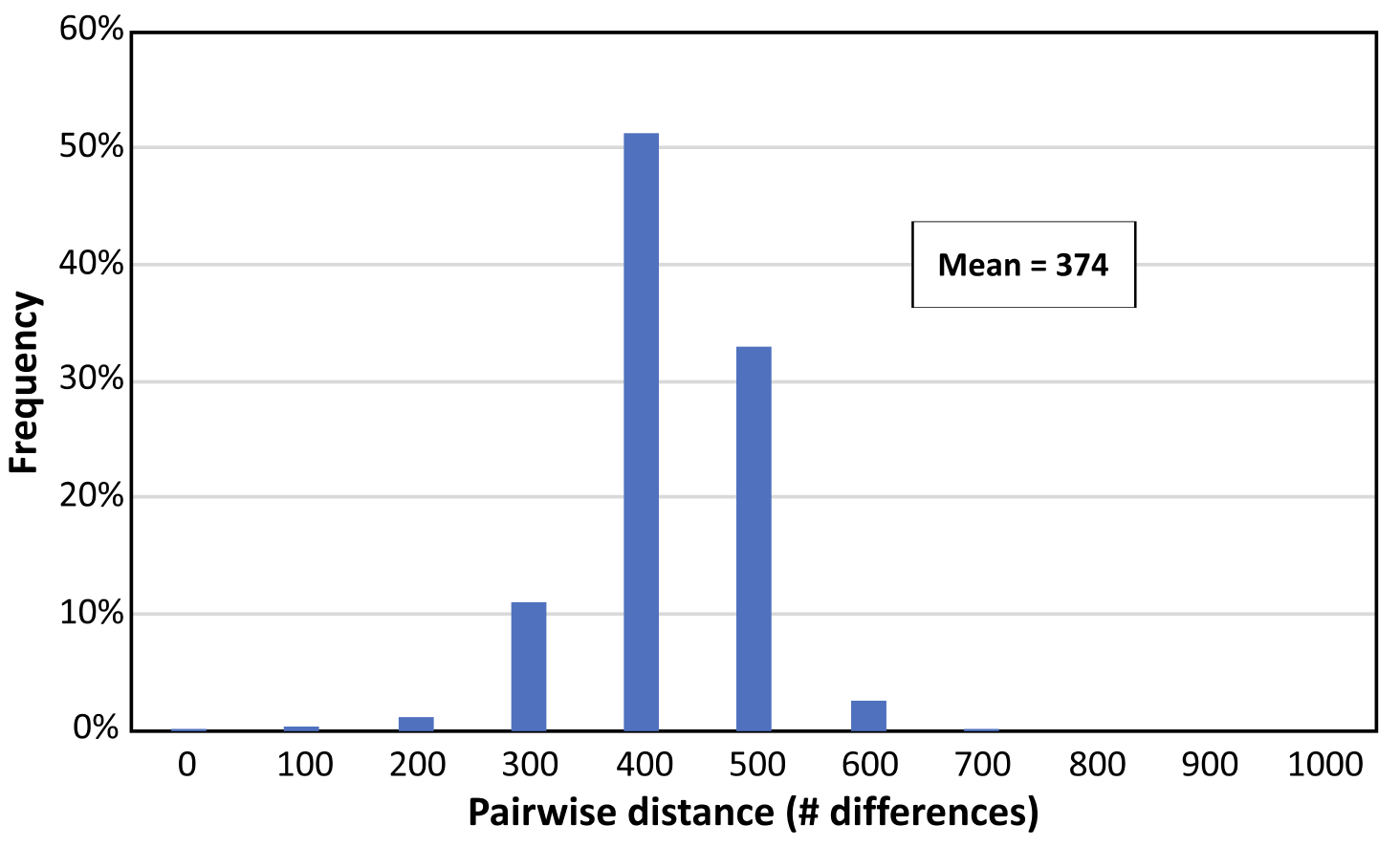
Figure 2. Distribution of the number of polymorphic markers between combinations of elite indica germplasm. Pairwise differences were based on the RiCA v.4 panel.
For more detailed information please consult https://isl.irri.org/services/genotyping/rica
Use in breeding
This panel can be used for genomic selection, QTL profiling, Background selection, Marker-assisted forward breeding, and QTL mapping.
References
Arbelaez J.D., Dwiyanti M.S., Tandayu E., Llantada K., Jarana A., Ignacio J.C., Platten J.D., Cobb J., Rutkoski J.E., Thomson M.J., Kretzschmar T. (2019) 1k-RiCA (1k-Rice Custom Amplicon) a novel genotyping amplicon-based SNP assay for genetics and breeding applications in rice. Rice 12:55

