Sweet potato mid-density genotyping services
Genotyping panel containing 3,210 SNP markers distributed evenly throughout the 15 chromosomes of sweet potato (Ipomoea batatas L.). This panel is suitable for marker-assisted breeding, linkage and introgression mapping, QTL analyses, outlier removal, and germplasm management applications.
The sweet potato DARTag_BI_Cornell_University (1.0) was developed from whole genome skim sequencing data from a diverse set of 47 sweet potato lines from 7 regional groups (Africa, Caribbean, Far East, Pacific islands, North America, Central America, and South America). Two biological replicates of each sample in the discovery panel were processed, where the sequencing libraries were prepared using either Illumina Nextera WGS library prep at the Genomics Facility of Cornell Institute of Biotechnology or NEBNext Ultra DNA Library Prep Kit at Novogene. Whole-genome sequencing with 150 bp paired-end reads was generated using Illumina NovaSeq 6000 at Novogene (https://en.novogene.com).
A total of ~41 M (41,625,649) SNPs were discovered from the whole-genome re-sequencing of the diversity panel, and where a high-confidence set of ~20K SNPs were obtained1,2,3. In addition to the previous filters, the 3120 SNPs were primarily selected from those with even genome distribution and locations in genic regions (Figure 1A). The sweet potato 3K DArTag marker panel was evaluated on a diverse set of elite lines of cultivated sweet potato (n=264) to assess the panel’s effectiveness on wider germplasm than was represented in the SNP discovery panel as shown by PCA (Figure 1B).
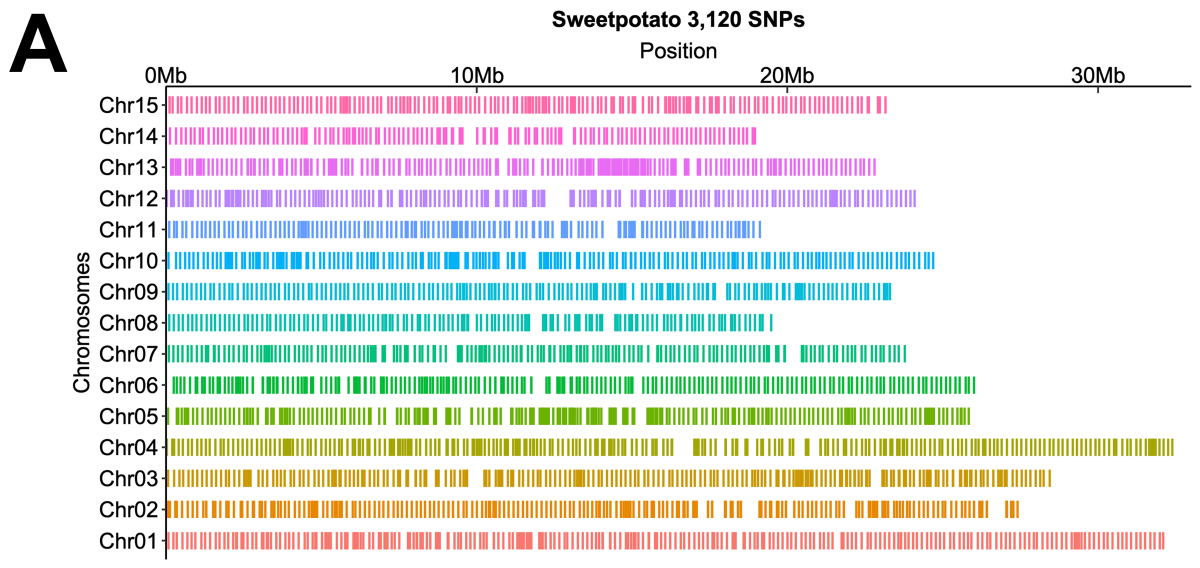

Figure 1. A) Selection of SNPs on the 3K panel as distributed by physical chromosomes. B) Three-dimensional PCA plot of the germplasm materials used to test the panel.
In addition to diversity testing, the panel was evaluated for linkage mapping ability with a reciprocal bi-parental population between ‘Beauregard’ and ‘Regal’ (36 and 56 progeny, respectively), a bi-parental population between ‘Beauregard’ and ‘Uplifter’ (20 progeny), the parents of which are from North America and Caribbean origin. After marker filtering a set of 1,476 high-quality SNPs were placed on the linkage Map (Figure 2A). The linkage map by Mollinari et al. 20204 was used as the framework, and the rearrangements observed in our study were anticipated and agreed with the marker order delineated in the 2020 map (Figure 2B).
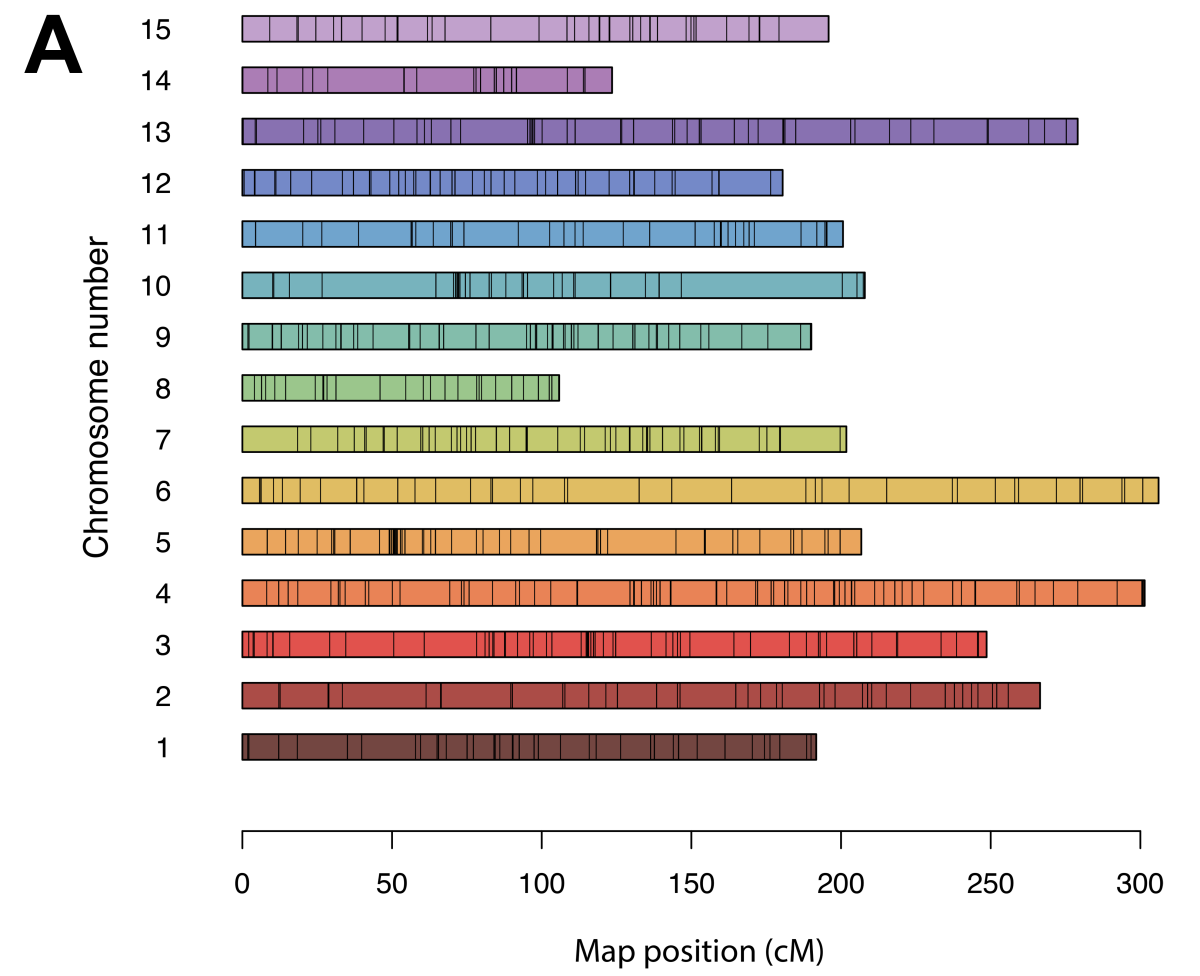
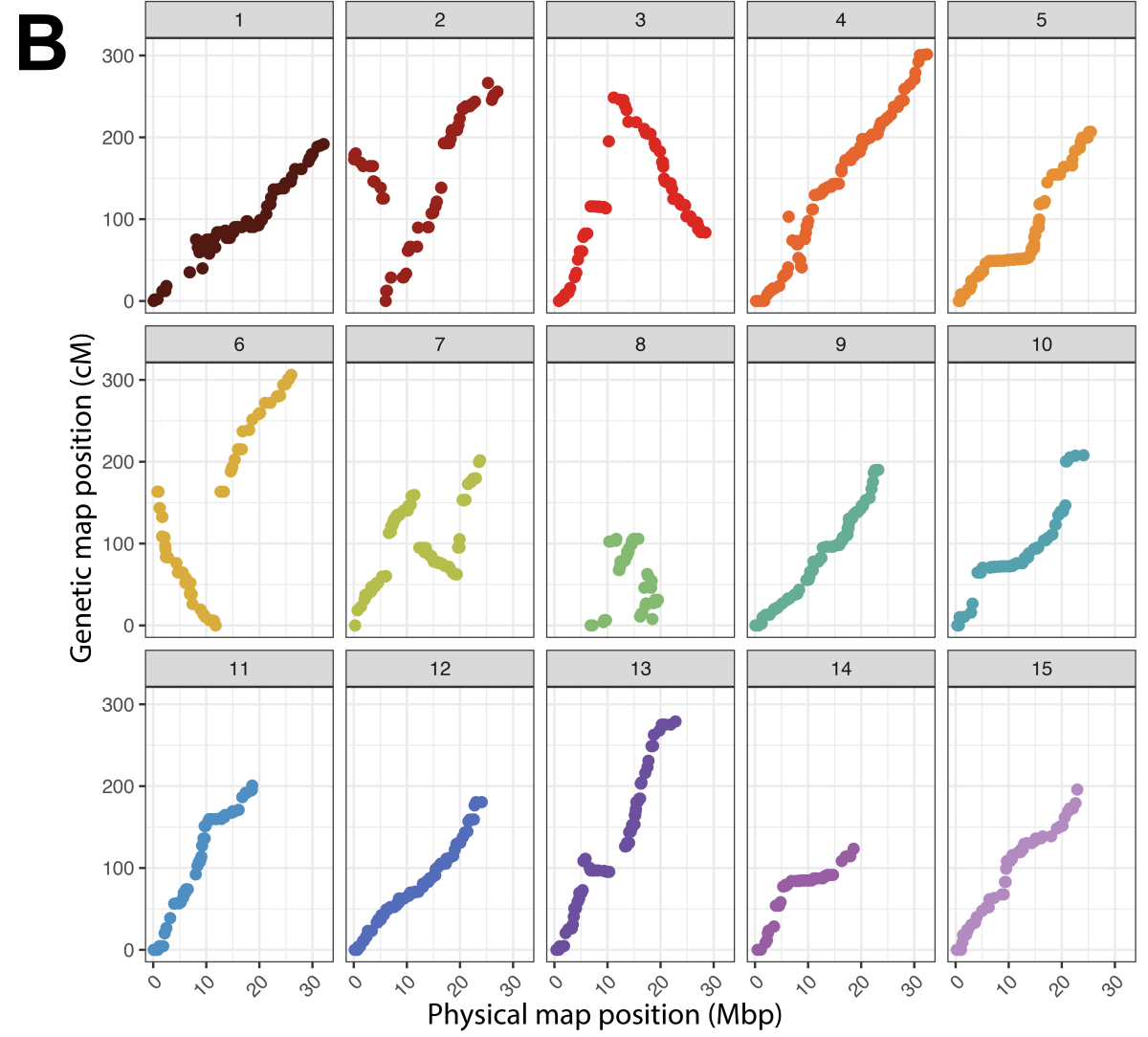
Figure 2. A) Chromosomal distribution of the 1,476 SNPs placed on a composite linkage map in genetic distance (cM). B) Correlation of the physical map (Mbp) versus the genetic map (cM) for each chromosome.
References
- Zhao et al. (2024). A public mid-density genotyping platform for sweetpotato (Ipomoea batatas), Genes (in review)
- Bolger, et al. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data, Bioinformatics, 30(15), pp. 2114–2120. doi: 1093/bioinformatics/btu170.
- Wu et al. (2018). Genome sequences of two diploid wild relatives of cultivated sweetpotato reveal targets for genetic improvement, Nature Communications, 9(1). doi: 1038/s41467-018-06983-8.
- Mollinari et al. (2020). Unraveling the Hexaploid Sweetpotato Inheritance Using Ultra-Dense Multilocus Mapping, G3 Genes|Genomes|Genetics, 10(1), 281–292. doi: 1534/g3.119.400620.
Results & data sharing
Breeding Insight invites panel users to share their MADC and sample metadata files by emailing them to bi-genotyping@cornell.edu to be added to the public microhaplotype database for fixed allele naming and public data sharing. Proprietary sample metadata can be privatized in the database for IP protection, upon request.
Disclaimer
These materials are based upon work supported by the U.S. Department of Agriculture, under agreement numbers [8062-21000-043-004-A, 8062-21000-052-002-A, and 8062-21000-052-003-A). Any opinions, findings, conclusions, or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the views of the U.S. Department of Agriculture. In addition, any reference to specific brands or types of products or services does not constitute or imply an endorsement by the U.S. Department of Agriculture for those products or services.

