The maize mid-density panel is an SNP panel developed and optimized by the collaboration between the International Maize and Wheat Improvement Center (CIMMYT) and the International Institute of Tropical Agriculture (IITA) to implement genomic assisted breeding routinely in the maize breeding programs.
This robust, cost-effective mid density panel was recently updated and currently consists of 3305 genome wide SNP markers and developed from sequencing data of >10,000 breeding lines and landraces belonging to different breeding programs from Mexico, Latin America, Africa and Asia for biotic and abiotic stress tolerance, adaptation and agronomic traits.
The sequence information for the development of the current 3.3K SNP panel are mainly derived from the genomics resources from CIMMYT and IITA including whole-genome re-sequencing (WGS), genotyping by sequencing (GBS), DArTseq genotyping, maize haplotype vs 3 (HapMap3) and Kompetitive Allele Specific PCR (KASP) SNPs (Bukowski et al., 2018; Chen et al., 2016; Gowda et al. 2017; Semagn et al., 2014; Wu et al., 2016).
The set expresses the several discovery studies made in maize from the last twenty years and bring together trait linked markers along with random markers spread across the genome, including the 70 QC KASP markers that are routinely used for progeny purity and F1 hybridity testing and described in the low-density KASP marker platform (https://excellenceinbreeding.org/module3/kasp).
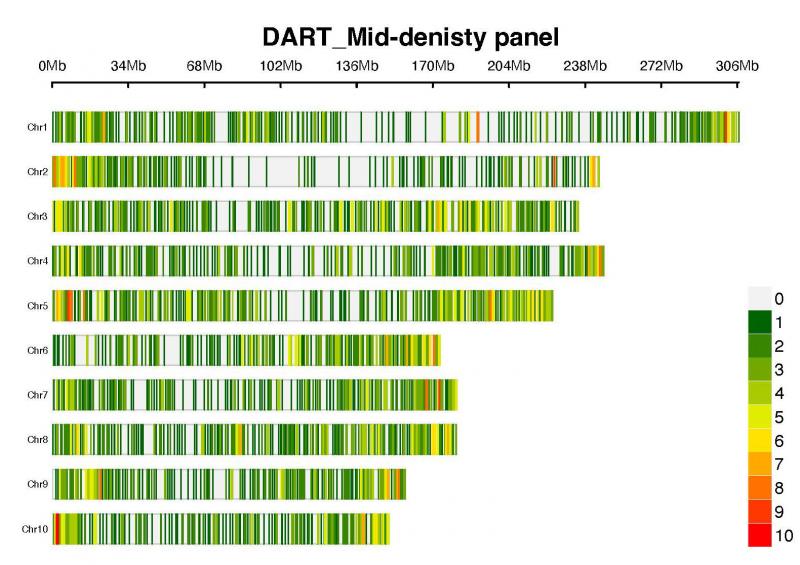
The average marker density of the panel is about 1 SNP per 0.72 Mbp, distributed across 10 chromosomes (Figure 1).
A comparison of prediction correlations between different maize SNP panels, namely DArTtag and rAmpSeq panels, in maize and applied to genomic predictions showed similar accuracy for grain yield and other traits under both optimum and drought management. These results support the suitability of this panel for GS applications in maize.
The CIMMYT and IITA Maize Programs currently use the 3.3K SNP panel to accelerate their breeding schemes for increased overall genetic gains through the ‘Accelerating Genetic Gains in Maize and Wheat’ and ‘Accelerated Breeding initiative’ projects funded by the BMGF, USAID, FFAR and banner of CtEH donors.
References
- Bukowski R, Guo X, Lu Y, et al. (2018) Construction of the third-generation Zea mays haplotype map, Gigascience, 7(4):1-12. https://doi.org/10.1093/gigascience/gix134
- Chen J, Zavala C, Ortega N, et al. (2016) The development of quality control genotyping approaches: a case study using elite maize lines, PLoS One, 11(6): e0157236. https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0157236
- Gowda M, Worku M, Nair, S.K, et al. (2017) Quality Assurance/ Quality Control (QA/QC) in maize breeding and seed production: theory and practice, http://hdl.handle.net/10883/19046
- Semagn K, Babu R, Hearne S, et al. (2014) Single nucleotide polymorphism genotyping using Kompetitive Allele Specific PCR (KASP): overview of the technology and its application in crop improvement, Molecular Breeding, 33: 1–14. https://doi.org/10.1007/s11032-013-9917-x
- Wu Y, San Vicente F, Huang·K, et al. (2016) Molecular characterization of CIMMYT maize inbred lines with genotyping-by-sequencing SNPs, Theoretical and Applied Genetics, 129:753–765. https://doi.org/10.1007/s00122-016-2664-8

