S. Dreisigacker, S. Esposito, Fillipo Bassi, Paolo Vitale, K. Ammar, and Pasquale de Vita
The 4K TdDArTAG panel v.01 is a mid-density SNP genotyping panel for durum wheat, developed at the International Maize and Wheat Improvement Center (CIMMYT, Mexico) in close collaboration with the Council for Agricultural Research and Economics (CREA, Italy) and the International Center for Agricultural Research in the Dry Areas (ICARDA, Morocco). The panel is designed to provide an affordable and efficient tool for genotyping in diversity analyses, quality control/quality assurance (QC/QA), and genomic selection programs.
This first version of the durum wheat panel includes 4,031 SNP markers distributed across the 14 durum wheat chromosomes (Figure 1; Table 1).
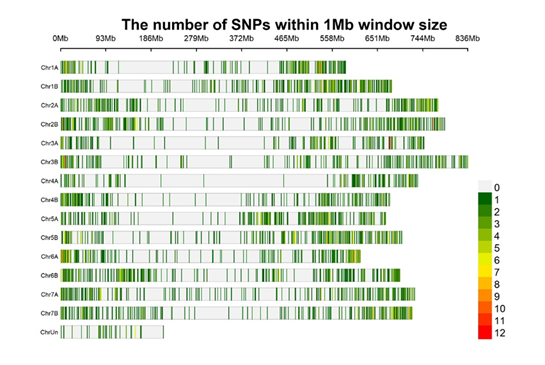
Figure 1. SNP density plot for 3392 SNPs in the current Wheat TdDArTAG panel vs. 01 aligned to the Svevo reference genomes vs. 1 (Source: Susanne Dreisigacker)
The sequences included in this panel are primarily derived from CREA and ICARDA genomic resources, based on large genotypic datasets generated using wheat SNP microarrays (9K and 90K iSelect arrays, Wang et al 2014). In addition, a set of 374 CIMMYT elite lines was genotyped using the bread wheat 3.9K DArTAG panel (TaDArTAG v.2), and the SNPs found to be polymorphic in durum wheat were incorporated into this panel. This contributed 989 overlapping SNPs (Table 2).
Table 1. Distribution of SNPs across genomes and chromosomes of durum wheat
| Chromosome → Genome ↓ |
1 | 2 | 3 | 4 | 5 | 6 | 7 | Un | Total Genome |
| A | 254 | 332 | 202 | 177 | 284 | 278 | 290 | – | 1817 |
| B | 366 | 357 | 270 | 217 | 339 | 316 | 305 | – | 2170 |
| Total Chromosome | 620 | 689 | 472 | 394 | 623 | 594 | 595 | 44 | 4031 |
The SNP markers were selected based on their overall polymorphism, minor allele frequency, genome and chromosome allocation, and overall genome distribution. A total of 159 consensus sequences for markers associated with several key genes/traits, i.e., disease resistance, quality, and agronomic traits, as well as markers linked to QTL, were included. Low-density genotyping for some of these trait-based markers is utilized in CIMMYT wheat breeding programs (see KASP marker list).
A detailed list of markers is available here.
Table 2. Number of sequences selected for the TdDArTAG panel v.01
| Type | # of sequences |
| Gene-based SNPs/Indels | 159 |
| QTL- associated SNPs | 446 |
| Genome-wide SNPs | 3426 |
| Overlapping with the TaDArTAG panel vs. 2 | 989 |
| Total | 4031 |
References
- Wang S., Wong D., Forrest K., Allen A., Chao S., et al., 2014. Characterization of polyploid wheat genomic diversity using a high-density 90,000 single-nucleotide polymorphism array. Plant Biotechnol. J. 12: 787–796. 10.1111/pbi.12183.

