Within the “Accelerating Genetic Gains in Maize and Wheat” and “Zinc mainstreaming in Wheat” projects funded by the BMGF, the UK FCDO, the USAID, and the FFAR; CIMMYT is piloting to further shortening its breeding cycle to increase its breeding efficiency. A robust, cost-effective mid-density SNP platform is ideal for this type of genomic-assisted breeding approach.
The wheat DArTag SNP panel vs. 1 currently contains 2,417 SNP markers distributed across the 21 bread wheat chromosomes (Figure 1). Sequences were derived from a large number of genomic resources at CIMMYT, including genotyping-by-sequencing data for more than 60K CIMMYT elite breeding lines and SNP array data on more than 1000 diverse wheat accessions or lines (Sehgal et al. 2020, Velu et al. 2016).
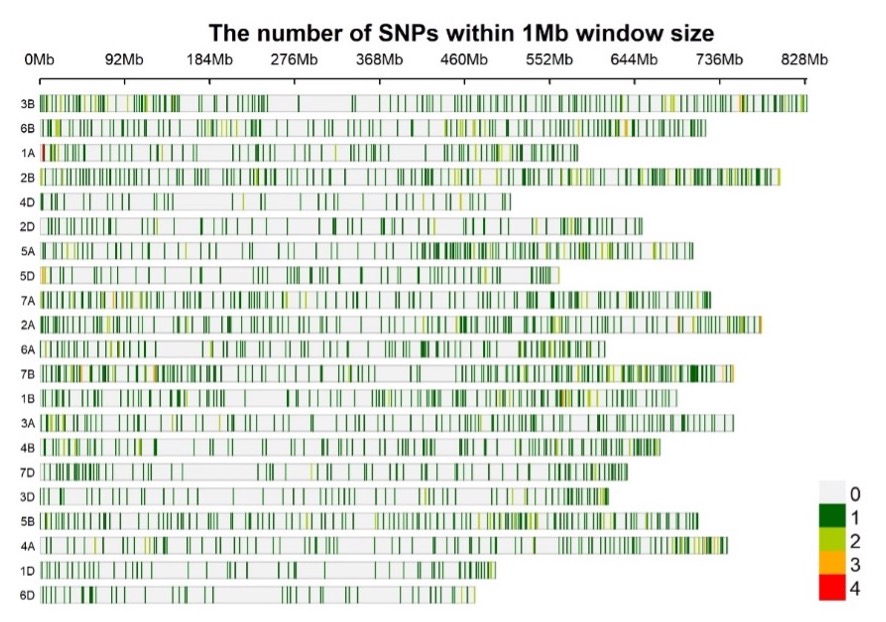 Figure 1. SNP density plot for 2417 SNPs in the Wheat DArTag panel vs. 01 (Source: Susanne Dreisigacker)
Figure 1. SNP density plot for 2417 SNPs in the Wheat DArTag panel vs. 01 (Source: Susanne Dreisigacker)
The SNP markers were selected based on their minor allele frequency, haplotypes formed in CIMMYT elite germplasm and an appropriate overall genome distribution. The several selected SNPs are related to target genes and identified QTL, in addition to a subset of around 480 sequences derived from exome capture sequencing of 890 diverse accessions of hexaploid and tetraploid wheat (He et al. 2019).
A phase 2 design is currently ongoing to add additional SNPs and planned to be available by mid of 2021.
This panel can be used for diversity studies and genomic selection.
Table 1. Number of sequences selected for DArTAG panels
| Type | # of sequences |
| Gene-based SNPs/Indels | 156 |
| QTL- associated SNPs | 312 |
| QC candidate SNPs | 110 |
| Genome-wide SNPs | 1802 |
| - GBS derived | 1280 |
| - 90K iSelect array derived | 522 |
| Exome capture SNPs (KSU) | 500 |
| UK35K Axiom Array SNPs (Bristol) | 1005 |
| Total | 3900 |
References:
- He, F., Pasam, R., Shi, F. et al. (2019). Exome sequencing highlights the role of wild-relative introgression in shaping the adaptive landscape of the wheat genome. Nat. Genet 51, 896–904. https://doi.org/10.1038/s41588-019-0382-2.
- Sehgal, D., Mondal, S., Crespo-Herrera, L. et al. (2020). Haplotype-Based, Genome-Wide Association Study Reveals Stable Genomic Regions for Grain Yield in CIMMYT Spring Bread Wheat. Front. Genet., 11, 1427. http://doi.rog/10.3389/fgene.2020.589490.
- Velu, G., Crossa, J., Singh, R.P. et al. (2016). Genomic prediction for grain zinc and iron concentrations in spring wheat. Theor. Appl. Genet. 129(8), 1595-1605. https://doi.org/10.1007/s00122-016-2726-y.

